Authors: Hayes H, Sullivan D, Millard D, and Clements M.
Axion BioSystems, Atlanta, GA
SfN 2021 Presentation
Multiwell MEA Technology
Microelectrode array technology
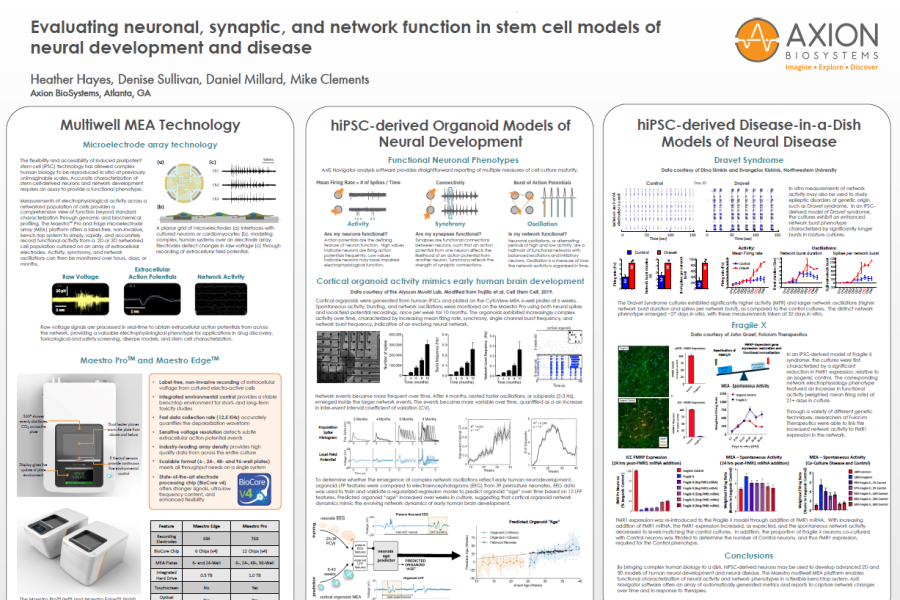
The flexibility and accessibility of induced pluripotent stem cell (iPSC) technology has allowed complex human biology to be reproduced in vitro at previously unimaginable scales. Accurate characterization of stem cell derived neurons and network development requires an assay to provide a functional phenotype.
Measurements of electrophysiological activity across a networked population of cells provides a comprehensive view of function beyond standard characterization through genomic and biochemical profiling. The Maestro ™Pro and Edge microelectrode array (MEA) platform offers a label free, non invasive, bench top system to simply, rapidly, and accurately record functional activity from a 2D or 3D networked cell population cultured on an array of extracellular electrodes. Activity, synchrony, and network oscillations can then be monitored over hours, days, or months.
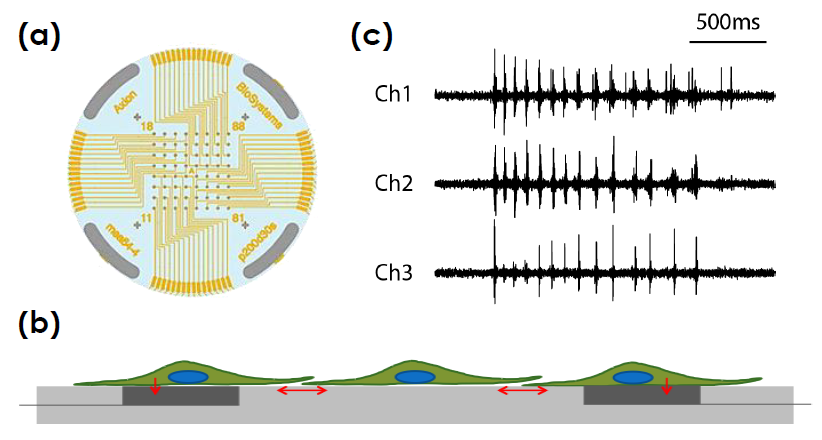
toxicological and safety screening, disease models, and stem cell characterization.
Maestro ProTM and Maestro EdgeTM
- Label free, non invasive recording of extracellular voltage from cultured electro active cells
- Integrated environmental control provides a stable benchtop environment for short and long term toxicity studies
- Fast data collection rate (12.5 KHz) accurately quantifies the depolarization waveform
- Sensitive voltage resolution detects subtle extracellular action potential events
- Industry leading array density provides high quality data from across the entire culture
- Scalable format (6-, 24-, 48- and 96- well plates) meets all throughput needs on a single system
- State of the art electrode processing chip (BioCore v4) offers stronger signals, ultra low frequency content, and enhanced flexibility

| Feature | Maestro Edge |
Maestro Pro |
|---|---|---|
| Recording Electrodes | 384 | 768 |
| BioCore Chip | 6 Chips (v4) | 12 Chips (v4) |
| MEA Plates | 6- and 24-Well | 6-, 24-, 48-, 96-Well |
| Integrated Hard Drive | 0.5 TB | 1.0 TB |
| Touchscreen | No | Yes |
| Optical Stimulation | Yes | Yes |
hiPSC derived Organoid Models of Neural Development
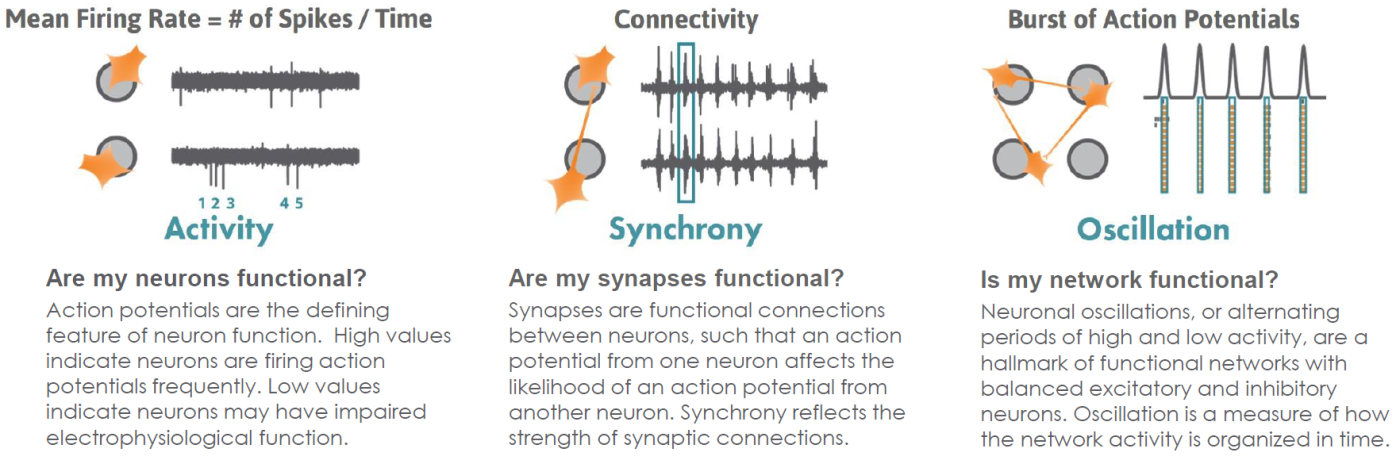
Functional Neuronal Phenotypes
AxIS Navigator analysis software provides straightforward reporting of multiple measures of cell culture maturity.
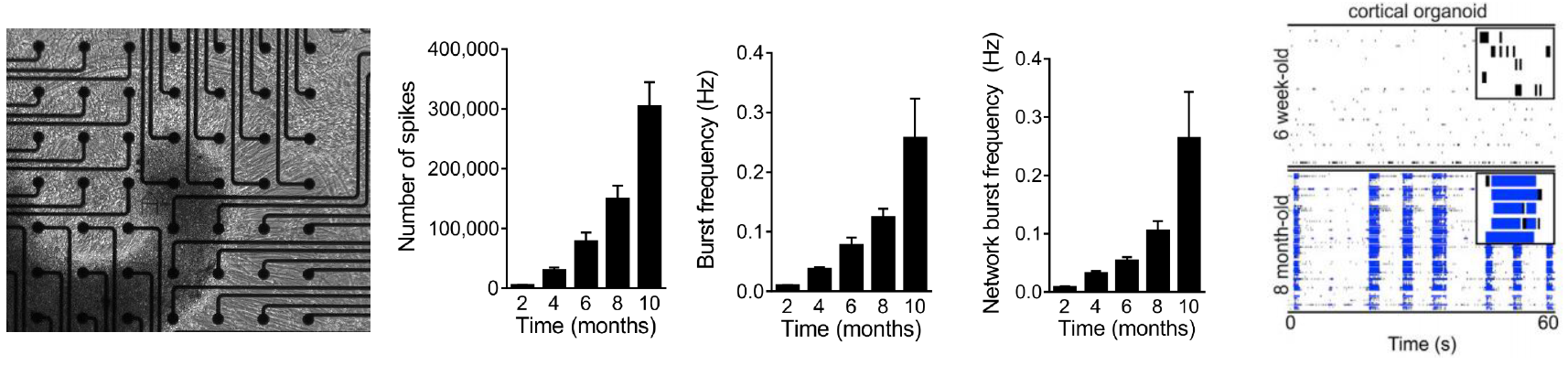
Cortical organoid activity mimics early human brain development
Data courtesy of the Alysson Muotri Lab. Modified from Trujillo et al, Cell Stem Cell, 2019.
Cortical organoids were generated from human iPSCs and plated on the CytoView MEA 6 well plates at 6 weeks. Spontaneous activity, bursting, and network oscillations were monitored on the Maestro Pro using both neural spikes and local field potential recordings, once per week for 10 months. The organoids exhibited increasingly complex
activity over time, characterized by increasing mean firing rate, synchrony, single channel burst frequency, and
network burst frequency, indicative of an evolving neural network.
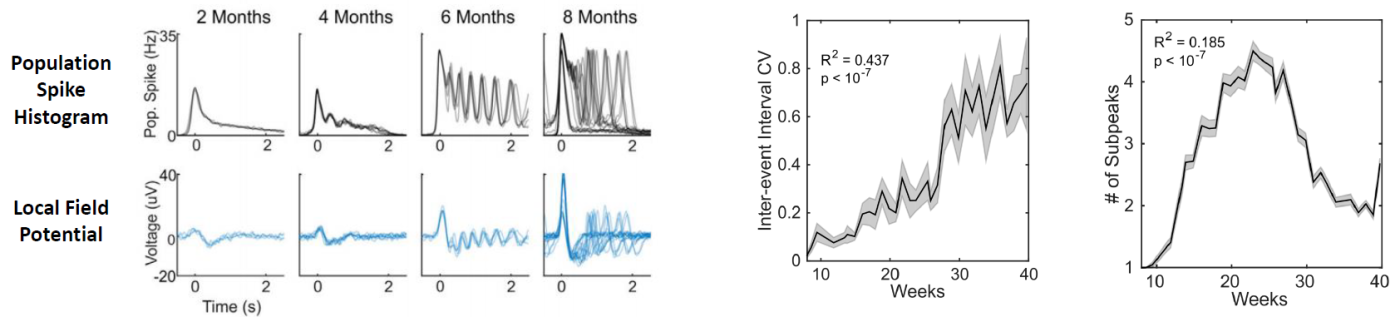
in inter event interval coefficient of variation (CV).
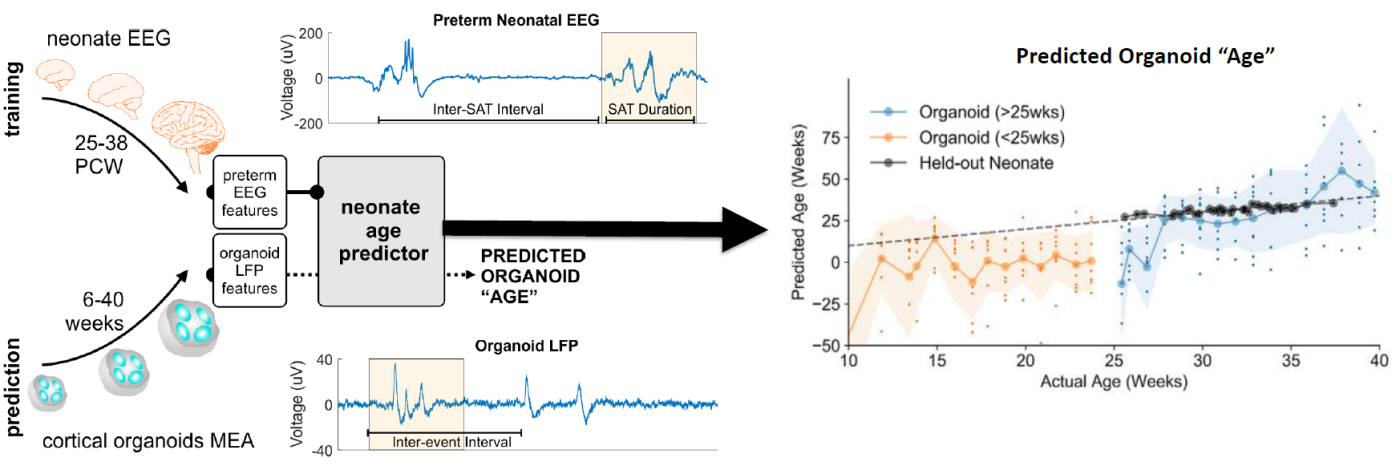
hiPSC-derived Disease in a Dish Models of Neural Disease
Dravet Syndrome
Data courtesy of Dina Simkin and Evangelos Kiskinis, Northwestern University
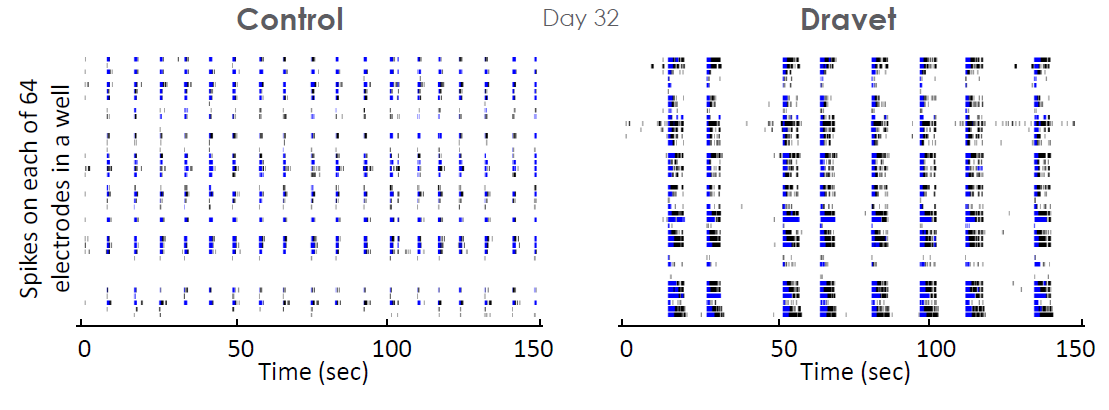
In vitro measurements of network activity may also be used to study epileptic disorders of genetic origin, such as Dravet syndrome. In an iPSC derived model of Dravet syndrome, the cultures exhibit an enhanced network burst phenotype characterized by significantly longer bursts in mature cultures.
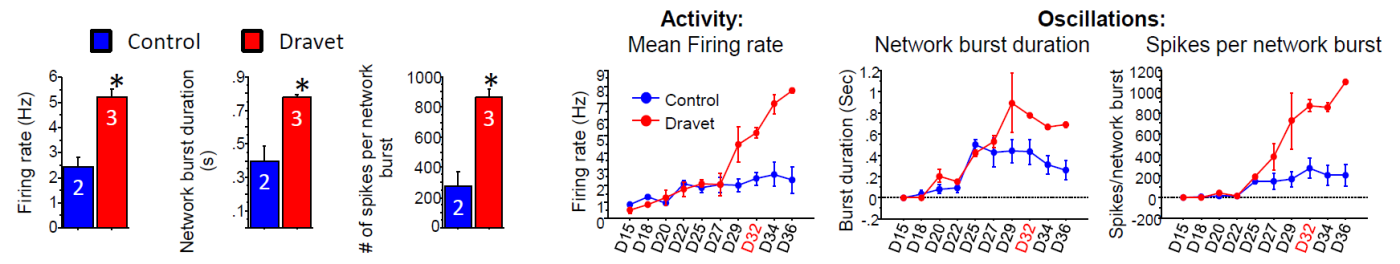
The Dravet Syndrome cultures exhibited significantly higher activity (MFR) and larger network oscillations (higher network burst duration and spikes per network burst), as compared to the control cultures. The distinct network phenotype emerged ~27 days in vitro , with these measurements taken at 32 days in vitro.
Fragile X
Data courtesy of John Graef, Fulcrum Therapeutics
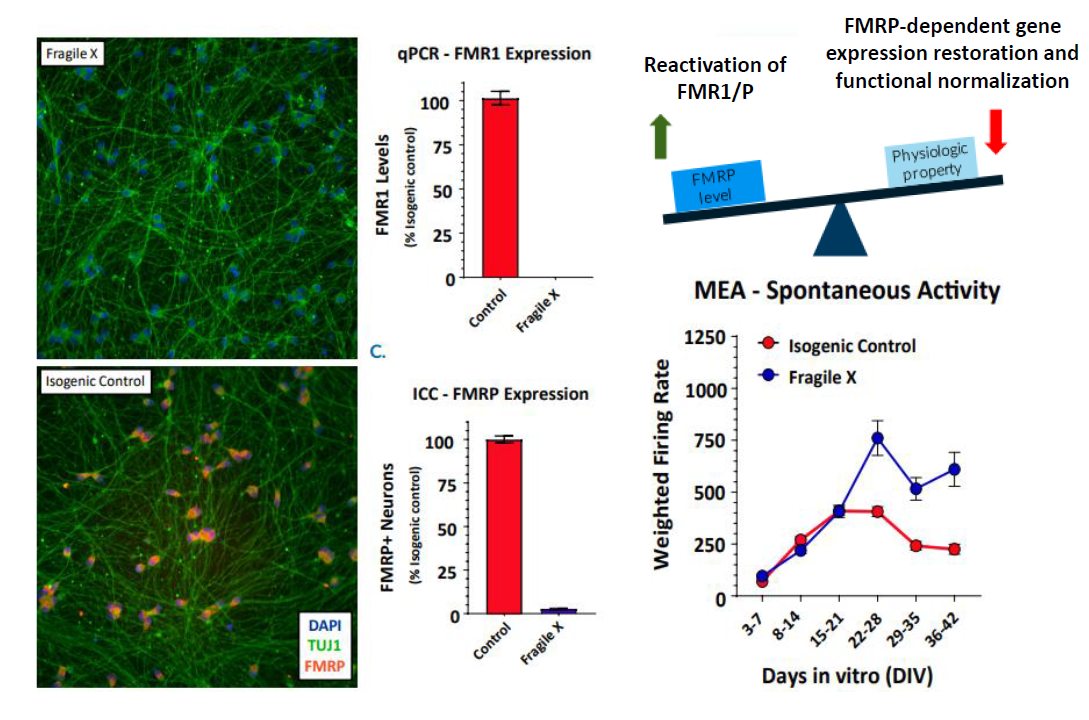
In an iPSC derived model of Fragile X syndrome, the cultures were first characterized by a significant reduction in FMR1 expression, relative to an isogenic control. The corresponding network electrophysiology phenotype featured an increase in functional activity (weighted mean firing rate) at 21+ days in culture. Through a variety of different genetic techniques, researchers at Fulcrum Therapeutics were able to link the increased network activity to FMR1 expression in the network.
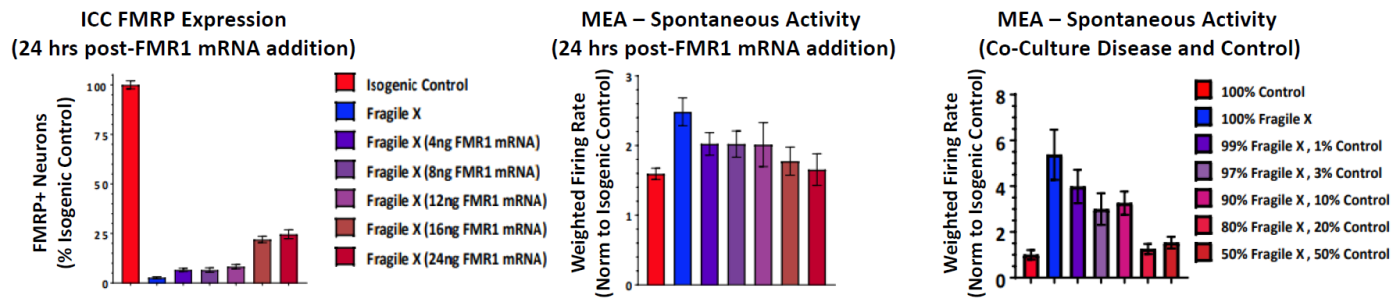
FMR1 expression was re-introduced to the Fragile X model through addition of FMR1 mRNA. With increasing addition of FMR1 mRNA, the FMR1 expression increased, as expected, and the spontaneous network activity decreased to levels matching the control cultures. In addition, the proportion of Fragile X neurons co cultured with Control neurons was titrated to determine the number of Control neurons, and thus FMR1 expression, required for the Control phenotype.
Conclusions
By bringing complex human biology to a dish, hiPSC derived neurons may be used to develop advanced 2D and 3D models of human neural development and neural disease. The Maestro multiwell MEA platform enables functional characterization of neural activity and network phenotypes in a flexible benchtop system. AxIS Navigator software offers an array of automatically generated metrics and reports to capture network changes over time and in response to therapies.