Key Findings
>> Pluripotent makers can be labeled and tracked with fluorescent imaging
Abstract
Maintenance and expansion of stem cell culture requires strict monitoring to ensure they retain their pluripotent or multipotent state for downstream applications. For example, pluripotent stem cell (PSC) cultures maintain tight cell colonies that grow larger over time and without close monitoring can be susceptible to spontaneous differentiation. Wholewell imaging allows users to quickly assess stem cell health and determine appropriate passaging ratios/ times. Furthermore, fluorescent scanning and live-cell imaging dyes reveal expression of critical stem cell-associated surface markers.
Introduction
Due to their ability to spontaneously differentiate into other cell types, in vitro stem cell cultures require strict monitoring and attention to detail to maintain their pluripotent or multipotent state during growth and expansion phases prior to any downstream differentiation protocols. Pluripotent stem cells (PSCs) such as embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) are an invaluable cell source for tissue engineering and regenerative medicine therapies due to their ability to differentiate into all three germ layers (endoderm, mesoderm, and ectoderm). Stem cell differentiation into cardiomyocytes1, neurons2, endothelial cells3, pancreatic β-cells4, and more has already been achieved. Conventional microscopes are often used to monitor the status and health of these cultures, but this method is limited by small fields of view, which only allow for visualization of a small number of colonies at a time and user-to-user variability when estimating coverage of stem cell cultures. In contrast, automated stem cell monitoring reduces the time and effort needed to track and analyze stem cell cultures. The Omni is a live-cell imaging platform capable of continuous multiwell imaging directly from the incubator. Whole-well brightfield scans and fluorescent imaging acquisition can be used to streamline the stem cell workflow by providing an easy, automated method for users to monitor stem cell cultures over time, evaluate the expression of stem cell-associated proteins, minimize the time cells spend outside the incubator, and reduce tedious collection and analysis of images from conventional microscopy. Here, we present an example workflow to monitor iPSC cultures using the Omni platform (Fig. 1).
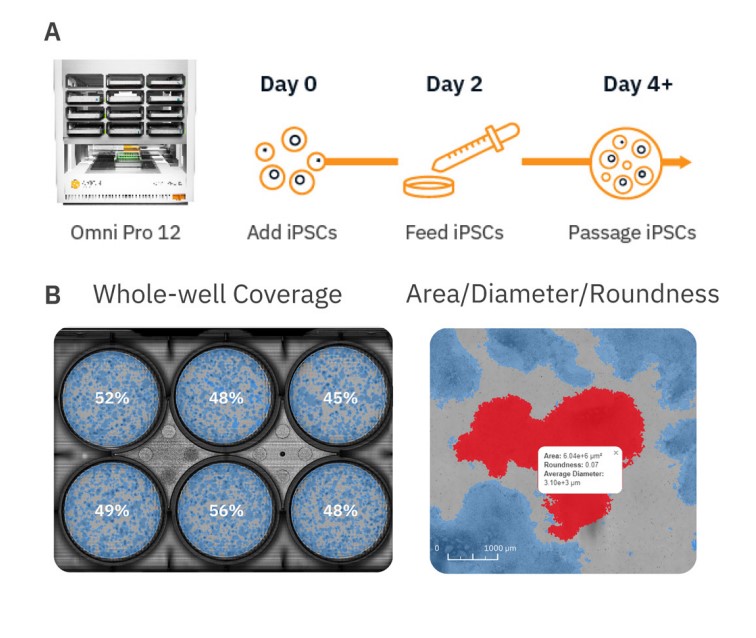
Materials and Methods
Cells and reagents
Human induced pluripotent stem cells (iPSCs; Cat. 200-0511) were obtained from Stem Cell Technologies (Vancouver, CA). Human iPSC media was composed of mTesR Plus (Stem Cell Technologies, Cat.100-0276). ESC-qualified Matrigel (Cat. 354277) was obtained from Corning (Corning, NY).
iPSC culture
First, Matrigel was used to coat the surfaces of 6-well tissue culture-treated plates (Corning, Cat. 38015). Matrigel aliquots (volume as specified by the certificate of analysis provided with each Matrigel lot) were thawed on ice and diluted in 25 mL of cold DMEM/F-12 medium (ThermoFisher, Cat. 11320033). Approximately 1 mL of diluted Matrigel was added to each well of a 6-well plate and incubated at room temp for 1 hour. Excess Matrigel was aspirated prior to iPSC plating.
Human iPSCs were thawed and cultured according to the supplier recommendations. Briefly, one vial of 1 million iPSCs was thawed and plated at different dilutions on Matrigel-coated 6-well plates. On Day 6-8 post thaw, the well with optimal iPSC colony density was selected for passaging. ReLeSR (Stem Cell Technologies, Cat. 05872) was used to lift iPSC colonies from the selected well and cells were split at a ratio of 1:30 into a new Matrigel-coated 6-well plate. Medium changes were performed every other day to maintain iPSC cultures.
Fluorescent staining
Human iPSC colonies were stained with pluripotent markers, TRA-1-60 and SSEA4-1, at Day 4 in culture. TRA-1-60 Alexa Fluor 488 Conjugate kit for Live Cell Imaging (ThermoFisher, Cat. A25618) and GloLIVE Human/Mouse SSEA-4 NL557 Live Cell Antibody (R&D Systems, Cat. NLLC1435R) were added directly to the iPSC culture at 1:50. After gentle swirling, the plate was incubated at 37ºC for 30 minutes. Then, the staining solution was aspirated and the cells were gently washed 2-3 times with FluoroBriteTM DMEM.
Omni imaging platform
The Omni platform was used to measure growth and coverage of iPSC cultures over time inside a cell culture incubator. During brightfield acquisition, the camera scans the sample stage and acquires a series of sequential images. These are stitched together to form an image of the total surface area of the culture vessel. When acquiring fluorescence images, users define how many snapshots within each well to record. All images are uploaded to the Axion Portal where they can be analyzed using AxIS Vue. For iPSC monitoring, the iPSC Module was used for analysis (Fig. 1).
Results
iPSC passaging confluence detection is streamlined with whole-well imaging
After the initial thaw of iPSC colonies, cells were plated at low to high densities in a 6-well Matrigel-coated plate. As described in the STEMCELL™ iPSC plating protocol5, each well received a different dilution of iPSCs. After 6 days of growth, the 6-well plate was imaged on the Omni to select the well with optimal iPSC colony density for passaging. Healthy iPSC cultures should consist of large, compact colonies with dense centers and well-defined borders. As shown in Figure 2, the Omni captures a brightfield whole-plate image of the iPSC culture for unbiased quantitative analysis. The coverage overlay (Fig. 2, top) provides a visual guide to the size and coverage of the iPSC colonies, while the quantitative assessment of iPSC coverage for each well facilitates decision making (Fig. 2, bottom). Whereas conventional microscopy techniques would require the user to scan the entire plate manually or use third-party software to estimate coverage, the Omni platform provides users with a streamlined and simple approach to determine the appropriate well to select for passaging based on their culture requirements.
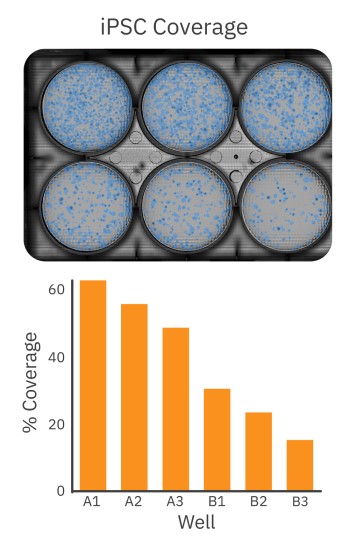
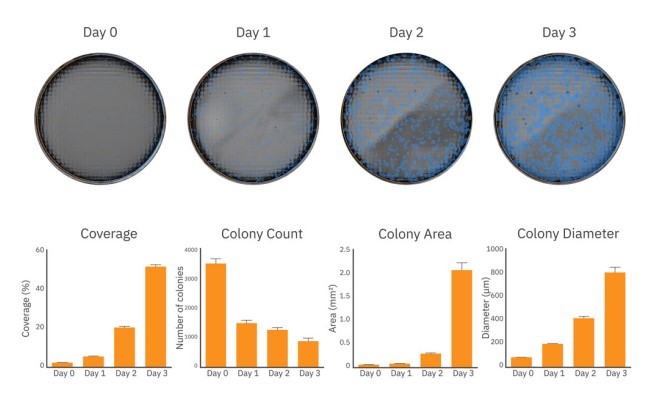
iPSC colony growth can be monitored and quantified over time
Passaging iPSC colonies at the ideal timepoint is critical to maintaining pluripotent and healthy colonies. Overgrown cultures may be more susceptible to spontaneous differentiation, thereby reducing the pluripotency of the overall cultures and negatively affecting the success of downstream differentiation protocols. Tracking culture density and colony size can help ensure consistent starting material from one differentiation to the next.
After passaging and plating, iPSC colony size and coverage in each well was monitored every day for 3 days on the Omni platform. As shown in Figure 3 (top), iPSC colonies grew in size and density from Day 0 to Day 3 of culture. By Day 3, iPSC growth was accelerating, as indicated by the rapid coverage and area increase from Day 2 (Figure 3, bottom), nearing the time to passage.
The coverage, colony count, average colony area and diameter at each timepoint provides the user with an accurate assessment for tracking iPSC growth.
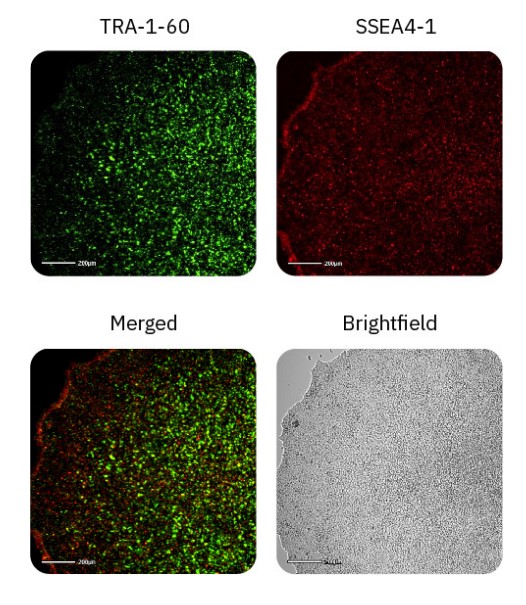
iPSC pluripotency can be detected with fluorescent imaging
Maintaining iPSC stemness (or pluripotency) is critical for successful iPSC cultures. The Omni platform not only allows for whole-plate visualization and calculation of iPSC coverage, but also allows for fluorescent imaging of important pluripotent markers that provide users information regarding the state of iPSC pluripotency.
TRA-1-60 and SSEA4-1 are well-known hallmarks of stem cell pluripotency and are expressed on the surface of ESCs and iPSCs6. These cell surface antigens are lost during the process of differentiation, and thus can be used as an indicator as to the state of pluripotency. Day 4 iPSC cultures were stained with TRA-1-60 Alexa Fluor 488 and SSEA4-1 NL577 and imaged in the green and red fluorescence channels on the Omni platform (Fig. 4). All iPSC cultures displayed TRA-1-60 and SSEA4-1 staining throughout the colonies, indicating that the iPSCs remained in a pluripotent state.
Conclusion
In this study, we demonstrated that iPSC cultures can be monitored and analyzed by the Omni platform to streamline the stem cell expansion workflow. Users can gain access to the growth and health of their stem cell cultures over time with automated, high-quality brightfield and fluorescent images.
References
[1.] Lian, X., Zhang, J., Azarin, S. et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/βcatenin signaling under fully defined conditions. Nat Protoc 8, 162–175 (2013).
[2.] Vieira MS, Santos AK, Vasconcellos R, Goulart VAM, Parreira RC, Kihara AH, Ulrich H, Resende RR. Neural stem cell differentiation into mature neurons: Mechanisms of regulation and biotechnological applications. Biotechnol Adv. 3, 7:1946-1970 (2018).
[3.] Jane Nguyen, Ying-Yu Lin, Sharon Gerecht, The next generation of endothelial differentiation: Tissue-specific ECs, Cell Stem Cell 28, 7: 1188-120. (2021).
[4.] Balboa, D., Barsby, T., Lithovius, V. et al. Functional, metabolic and transcriptional maturation of human pancreatic islets derived from stem cells. Nat Biotechnol 40: 1042–1055 (2022).
[5.] Healthy Control Human iPSC Line, Female, SCTi003-A. https://cdn.stemcell.com/media/files/pis/10000014504-PIS_03.pdf
[6.] International Stem Cell Initiative. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 25,7 (2007).
Authors
Benjamin Streeter, Denise Sullivan, Linda Boekestijn, Daniel Millard
Axion BioSystems, Atlanta, GA and Eindhoven, NL


