Parkinson’s disease (PD) is the second most common late-onset neurodegenerative disorder, clinically characterized by a series of motor symptoms such as bradykinesia, resting tremor, rigidity, and postural instability as a result of dopaminergic (DA) neuron degeneration in the midbrain. In an effort to dissect genetic vs non-genetic contributions to the disease, researchers from the New York Stem Cell Foundation embarked on a uniquely controlled study involving twins. Although both males carry the GBA N370S mutation known to increase Parkinson’s risk, only one twin has shown clinical manifestations of the disease. To study cellular and molecular differences between disease and non-disease states, researchers reprogrammed fibroblasts from each twin into induced pluripotent stem cells (iPSCs) and then differentiated the iPSCs into DA neurons. These derived neurons were then used in numerous in vitro assays, including activity analysis using the Maestro™ microelectrode array (MEA) platform.
Introduction
Parkinson’s disease (PD) is the second most common late-onset neurodegenerative disorder, clinically characterized by a series of motor symptoms such as bradykinesia, resting tremor, rigidity, and postural instability as a result of dopaminergic (DA) neuron degeneration in the midbrain (1).
In an effort to dissect genetic vs non-genetic contributions to the disease, researchers from the New York Stem Cell Foundation embarked on a uniquely controlled study involving twins. Although both males carry the GBA N370S mutation known to increase Parkinson’s risk, only one twin has shown clinical manifestations of the disease. To study cellular and molecular differences between disease and non-disease states, researchers reprogrammed fibroblasts from each twin into induced pluripotent stem cells (iPSCs) and then differentiated the iPSCs into DA neurons (2). These derived neurons were then used in numerous in vitro assays, including activity analysis using the Maestro™ microelectrode array (MEA) platform.
Results
After confirmation of neural lineage via gene/protein expression and ion channel functionality with patch clamp, iPSC-derived DA neurons from each twin were cultured on Maestro MEA plates in order to assess differences in functional activity and network formation. Since MEA is non-invasive, researchers were able to record activity from the same cultured cell population over time to monitor differences in neural activity.

Maestro MEA allowed researchers to analyze electrophysiological activity and assess neural network activity through the simultaneous data collection across all 64 electrodes in each well. This combination of insight enabled the discovery that PD neurons are delayed in development of both spontaneous activity and a mature neural phenotype as compared to non-PD neurons of the same age.
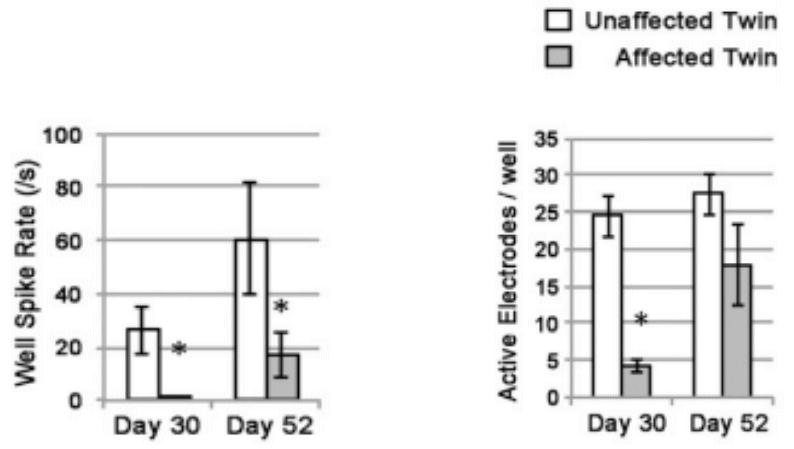
Benefits of Maestro MEA in PD disease modeling
- MEA allowed simultaneous in vitro examination of neural activity and connectivity from healthy and disease phenotypes using iPSC-derived neurons.
- MEA enabled long-term data collection from the same culture due to the non-invasive nature of extracellular voltage recording.
- Maestro MEA provided information from numerous locations within the cultured cell population, providing data on neural network connectivity and maturity.
References
- L.V. Kalia and A.E. Lang, Parkinson’s disease,” The Lancet, vol. 386, no.9996. pp. 896-912 (2015).
- Woodard et al., “iPSC-Derived dopamine neurons reveal differences between monozygotic twins discordant for Parkinson’s disease,” Cell Reports 9, 1173- 1182 (2014).



