This article is taken from European Biopharmaceutical Review October 2021, pages 36-40. © Samedan Ltd
Using induced pluripotent stem cells differentiated into neurons that recapitulate diseases, such as ALS and epilepsy, researchers are opening new doors for scientists to model and treat neurological diseases
The unique electrical properties of neurons make them challenging to study. Because of this, many cell biologists might feel that the brain is off limits – that only the most highly-trained electrophysiologists can unravel the mysteries of the nervous system. However, this is not the case. Thanks to advancements in bioelectroniconic assays, all cell biologists can now study neural networks without electrophysiology training.
Traditional electrophysiological techniques, such as patch-clamp recording require significant skill and hands-on time. Given the amount of training needed to perform this technique, the practice is almost exclusively performed by specialised electrophysiologists. However, even with experience and training, patch-clamp still has its limits. Researchers can only record data from a handful of cells per day. Furthermore, the technique enables recordings from only one neuron at a time and, therefore, does not capture the rich network activity that is characteristic of the brain. Finally, because neurons are so fickle ex vivo, a scientist also needs a bit of luck to ensure their experiment works out.
In contrast, bioelectronic assays, such as multielectrode array (MEA) assays, enable any researcher to simultaneously monitor the activity of dozens of cultured neural networks over time without specialised training. These assays are composed of a microtitre plate with grids of microelectrodes embedded in each well. These arrays are often used to record neural population activity on the surface of the brain in living animals; however, when they are embedded in a microtitre plate, they can make extracellular recordings of neural activity in vitro. This setup enables any cell biologist to analyse the neural activity of their cultures, track how networks develop, and monitor the emergence and progression of disease phenotypes at the benchtop. The accessibility of this technology adds a new tool to a neurobiologist’s toolbox to catalyse biomarker and drug discovery for neurological diseases.
The Challenge of Generating Cellular Models of Neurological Disease
The complexity of the brain has made it difficult for researchers to identify in vitro biomarkers of neurological disease that could assist in diagnosis and drug discovery. To advance the field, scientists must discover the functional underpinnings of disease, learn how they affect brain activity, and identify drugs that correct aberrant neural activity. This process starts by examining the impact of genetic and environmental manipulations on cellular models of disease.
Induced pluripotent stem cell (iPSC) technology has created an abundance of neuronal models that express the genetic profile of specific patients, allowing for in vitro testing of neurological conditions. A number of molecular methods exist to characterise these models. Measuring neuronal activity however, often requires low-throughput techniques, such as patch-clamp, that require extensive training and expertise. This creates a roadblock to testing, delaying progress and new discoveries.
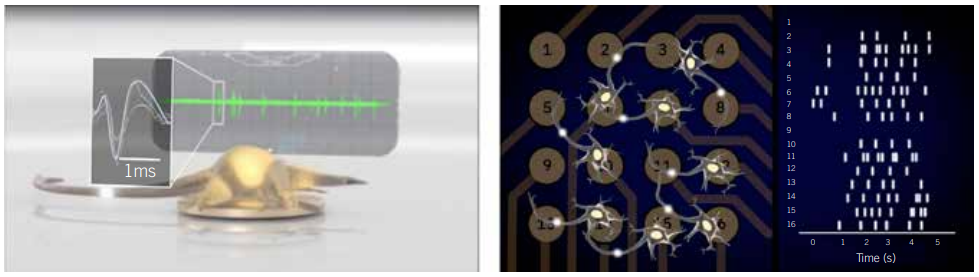
MEA assays remove that roadblock, bringing electrophysiological measurements to any biology lab. They enable cell biologists to create models of neurological disease in a dish, monitor the models’ electrophysiological properties, and learn more from their disease models than patch-clamp electrophysiology alone can offer. As evidenced by the examples in Figures 3 and 4 (page 38), this technology enables more biologists to study complex models of neurological and psychiatric conditions ranging from epilepsy to inherited erythromelalgia (IEM) in new ways on a lab bench.
A Precision Medicine Approach to Treating Epilepsy
Approximately 65 million people live with epilepsy. The condition comes in many forms, and, as a result, there is no one-size-fits-all approach to treatment. Yet, physicians do not have a test that can predict which treatment will work in each patient; instead, they must still rely on trial and error to find effective treatments. This approach fails for roughly one-third of patients (1). People with epilepsy could benefit from precision medicine, but researchers have not yet discovered the genetic biomarkers needed to enable this approach (2).
Evangelos Kiskinis PhD, Assistant Professor of Neurology at the Northwestern University Feinberg School of Medicine, US, is trying to solve this conundrum by developing an ‘epilepsy-in-a-dish’ model and probing it with an MEA assay. Many scientists model epilepsy using non-human cells and animals, but these models do not accurately reflect the unique characteristics of one’s condition, making it difficult to use these models to identify relevant biomarkers and discover effective treatments. Kiskinis, on the other hand, developed patient-specific models of epilepsy in vitro using iPSCs that he differentiated into neurons. Since the cells have the same genetic background as the patient they came from, the model better recapitulates the individual’s disease in a dish. Kiskinis is using these models to identify predictive biomarkers for epilepsy with the hope that, in the future, physicians will not have to rely on trial and error to identify effective treatments for their patients.
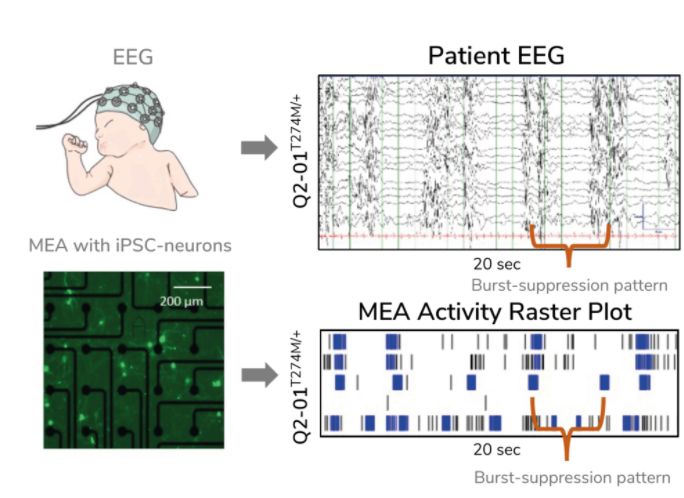
Kiskinis aimed to create a model of KCNQ2-associated epilepsy. There are more than 300 distinct genetic variations associated with this subtype of epilepsy, yet limited treatment options exist, creating a need for a more precise approach. Kiskinis and his team used an MEA assay to monitor iPSC-derived neuron activity overseveral weeks. He confirmed that his model reflected the characteristic burst-suppression firing pattern seen in KCNQ2-associated epilepsy (3). With a valid model of the disease, his team can test hundreds of variants to identify predictive biomarkers and identify more personal, targeted therapeutic options for patients.
A Biomarker of Treatment Effectiveness in Amyotrophic Lateral Sclerosis
Kiskinis and his team also used MEA assays to identify a potential treatment for amyotropic lateral sclerosis (ALS), a disease that causes the progressive loss of motor neuron function and paralysis. One in 50,000 people develop ALS, but there are no diagnostic markers or treatments for the disease (4). Scientists have identified over 20 ALS-associated genes, suggesting a role for precision medicine (5).
First, Kiskinis created an ‘ALS-in-a-dish’ model using patient-derived iPSCs harbouring the superoxide dismutase 1 alanine to valine mutation at codon 4 (SOD1A4V) mutation. Mutations in SOD1 are linked to about 15-20% of ALS cases, and the A4V mutation causes a rapidly progressing form of the disease. Using MEAs, he found that the cells were hyperexcitable, replicating previous studies that used manual patchclamp (6). Next, he corrected the mutation in the SOD1 gene and found that the correction rescued their normal function. This work demonstrated that neuronal excitability might be a useful biomarker of treatment effectiveness. Next, Kiskinis used his model to test the effect of an anti-epileptic drug called ezogabine that is known to reduce the excitability of neurons. He found that the drug reduced the excitability of motor neurons in the brain and spinal cord of ALS patients. His findings led the drug straight into a Phase II clinical trial (7).
A Precision Treatment for IEM
Like epilepsy and ALS, IEM, also called ‘burning man’ syndrome, is not easy to treat. The disease causes the sensation of intense, burning pain in one’s hands and feet, and traditional painkillers do not have any effect.
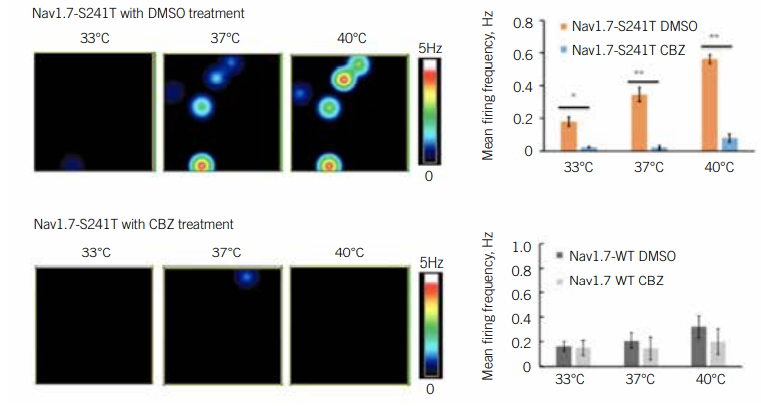
IEM is caused by a mutation in a gene that encodes a subunit of a sodium channel called NaV1.7. This channel mediates the activity of pain-sensing neurons. Scientists have discovered more than two dozen mutations that affect NaV1.7 function, indicating that no single treatment is likely to help all patients with the disease. To identify drugs that could help patients with IEM harbouring specific mutations, Yang Yang PhD, Assistant Professor of Medicinal Chemistry and Molecular Pharmacology at Purdue University, US, deployed an MEA assay.
Yang was interested in carbamazepine, a drug that physicians had discovered is effective against IEM in patients with the Nav1.7-V400M mutation. Yang wanted to find out if this drug worked in patients with other mutations (8). Yang first differentiated iPSCs with various mutations into pain-sensing neurons, called nociceptors. He then used an MEA assay to test the impact of carbamazepine on their activity. Since heat worsens IEM symptoms, he examined these neurons under several different temperature conditions. He identified a temperature-dependent increase in neuronal activity and firing frequency, as well as increased excitability overall, in mutant cells. Carbamazepine reduced this excitability of Nav1.7-V400M cells, but not in the wild type cultures. In a clinical setting, patients with this mutation reported reduced pain in response to the drug.
A Model of the Developing Brain
Conditions such as ASD and schizophrenia develop in utero, when the brain is still wiring itself. It is virtually impossible to study the developing human brain in this stage. Scientists have relied on animal models like mice and monkeys to provide insights into brain development, but these models are not the human brain.
Brain organoids generated from iPSCs mimic various developmental features at the cellular and molecular levels. Alysson Muotri PhD, Director of the Stem Cell Program at the University of California San Diego, US, has been working to characterise the development of brain organoids to assess their validity as a model of human brain development.
Muotri and his team plated cortical organoids in MEA plates. The assay generated activity maps and raster plots every week over 10 months, enabling them to study the changing electrophysiological activity of the model as it developed (9). During the experiment, the cortical organoids exhibited consistent increases in electrical activity. Also, channel-wise firing rate, burst frequency, and synchrony all shifted in a manner that indicated a continuously developing neural network. Overall, the organoid’s electrophysiological signature mimicked the preterm neonatal human brain, especially after 25 weeks post-conception.
Muotri’s iPSC-derived ‘mini-brains’ will help him understand the developing brain and test therapeutic interventions that could one day be used treat neurodevelopmental and psychiatric conditions, before it is too late.
The Role of Bioelectronic Assays in the Lab
Bioelectronic assays like MEA provide neuroscientists with a straightforward path to studying the electrical activity of neurons. They do not replace patch-clamp electrophysiology, but they open new doors for scientists to model neurological disease and ask different questions of their models. By studying these complex disease models using bioelectronic assays, researchers will be able to accelerate biomarker and drug discovery and enable neurologists to bring much-needed relief to more patients.
James Ross PhD is the Chief Technology Officer and Co-Founder of Axion BioSystems, as well as the Co-Founder and Scientific Advisor of BioCircuit. James has over 20 years’ experience in bioelectronic technology development, devoting much of his efforts to commercialising cost-effective cellular interfaces and software-reconfigurable analysis tools. He received his undergraduate degree in Electrical Engineering from Louisiana State University, US, in 2000, and his PhD in Neuroengineering from the Georgia Institute of Technology, US, in 2008.
References
1. Visit: www.sciencemag.org/news/2019/12/epilepsy-drugs-fail-nearly-one-third-patients-scientists-seek-root-causes-seizures
2. Ngugi AK et al, Estimation of the burden of active and life-time epilepsy: A meta-analytic approach, Epilepsia 51(5): pp883–90, 2010
3. Visit: elifesciences.org/articles/64434
4. Visit: www.als.org/navigating-als/resources/fyi-epidemiology-als-and-suspected-clusters
5. Chen S et al, Genetics of amyotrophic lateral sclerosis: an update, Mol Neurodegener 8(28), 2013
6. Wainger BJ et al, Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons, Cell Rep 7(1): pp1-11, 2014
7. Wainger BJ et al, Effect of ezogabine on cortical and spinal motor neuron excitability in amyotrophic lateral sclerosis, JAMA Neurol 78(2): pp186-96, 2021
8. Yang Y et al, NaV1.7 as a pharmacogenomic target for pain: moving toward precision medicine, Trends Pharmacol Sci 39(3): pp258-75, 2018
9. Trujillo CA et al, Complex oscillatory waves emerging from cortical organoids model early human brain network development, Cell Stem Cell 25(4): pp558-69.e7, 2019


